AI Hit Rates and Novelty
The AIDD Code Series
Part 1
Small-molecule drug discovery involves identifying chemicals that can interact with specific proteins and up- or down-regulate their activity to counteract a related disease state. Traditionally, this process relies on High-Throughput Screening (HTS) to test thousands of compounds in laboratory assays, with 2%, maximally, showing bioactivity, “hits”, against the target protein typically [1]. To improve compound hit rates, AI models are typically used early in the hit-finding process to screen compounds in silico before in vitro testing. AI drug discovery companies like Schrödinger and Insilico Medicine have made recent claims of hit rates of 26% [2] and 23% [3], respectively.
The lack of standardized hit rate reporting guidelines to evaluate the proficiency of different AI models confounds apples-to-apples comparisons between platforms, however. The calculation of hit rates may seem trivial at first glance, but they can vary drastically when considering variables such as chemical novelty, testing concentrations, and phase of hit discovery. Recognizing these complexities, our recent study [4] highlighted the performance of ChemPrint, developed by Model Medicines, which achieved a 46% hit rate, where 19 out of 41 AI-predicted compounds demonstrated novel biological activity in vitro. This blog aims to establish guidelines for evaluating AI drug discovery models, focusing on publications that provide real-world validation of predicted compounds.
“This progression from Hit Identification to Hit Optimization means that AI models typically perform better as the tasks shift from discovering novel chemistry to refining existing structures. ”
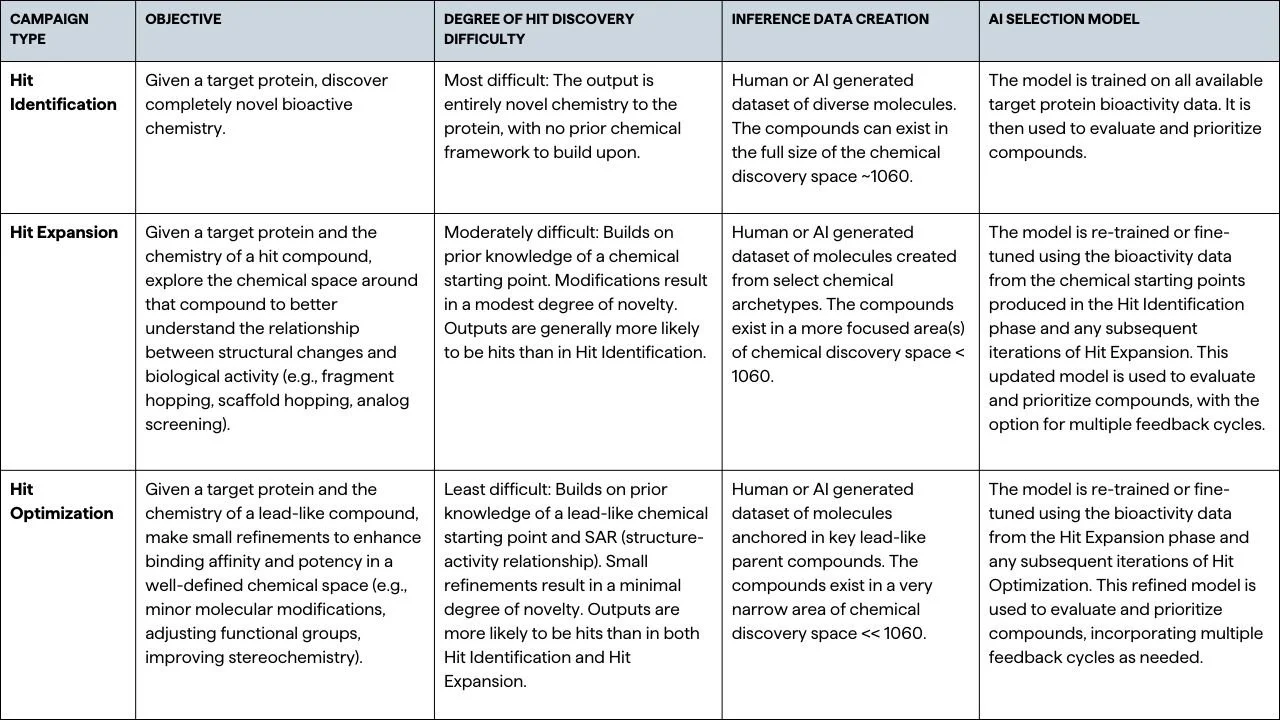
Different phases of drug discovery inherently have different hit rates, driven by the degree of prior knowledge and the novelty of the chemical space being explored, so it’s important to account for these differences when evaluating how well AI models perform. We classify the three major types of hit discovery campaigns as Hit Identification, Hit Expansion, and Hit Optimization, each representing a distinct phase in the discovery process (Table 1). Hit Identification is the most challenging phase, as it involves discovering entirely novel bioactive chemistry for a target protein. In contrast, Hit Expansion leverages a chemical starting point and explores specific areas in chemical space around a hit compound through modification techniques such as fragment hopping, scaffold hopping, or analog screening. This phase is moderately difficult, with outputs showing modest chemical novelty compared to the bioactive starting point, making them generally more likely to be hits than those in the prior Hit Identification phase. Hit Optimization is relatively the least challenging phase, involving minimal refinements to an already well-defined lead-like compound within a specific and well-understood Structure-Activity Relationship (SAR) framework. The chemical novelty of the outputs here is minimal, and the likelihood of discovering hits is significantly higher compared to the earlier phases. These inherent differences in hit rates, driven by the level of prior knowledge, carry over into AI-driven discovery as well. The more similar the predicted chemistry is to that of the training data, the easier it is for the AI model to predict molecular behavior accurately. This progression from Hit Identification to Hit Optimization means that AI models typically perform better as the tasks shift from discovering novel chemistry to refining existing structures. Understanding these nuances is crucial when comparing AI model performance, highlighting the importance of aligning evaluation metrics with the specific challenges of each campaign type.
In this blog post, we will evaluate in vitro validated AI hit rates from the literature specifically from within Hit Identification campaigns, where the challenge of discovering novel compounds is most pronounced.
Table 1.
Overview of campaign types in drug discovery, detailing objectives, difficulty levels, data creation methods, and AI models across Hit Identification, Hit Expansion, and Hit Optimization phases.
“...We believe the only thing that truly matters is the endpoint—can the AI platform discover the drug that solves the disease?”
In evaluating AI-driven small molecule drug discovery, the type of model used for compound selection is less important than two critical components for evaluation: the in vitro validation of AI-predicted compounds and the availability of the training data utilized. While it might seem unfair to compare different types of AI approaches, we believe the only thing that truly matters is the endpoint—can the AI platform discover the drug that solves the disease? This is the ultimate measure of success. With this in mind, we conducted a thorough analysis of the current landscape of AI models used for Hit Identification campaigns, focusing on publications that provide real-world validation of predicted compounds and breaking down the data behind their claimed hit rates. All data and analysis presented in this blog post are publicly available on our GitHub. We extend special thanks to the recent review article Machine Learning-Aided Generative Molecular Design [5], which consolidated numerous academic and commercial AI drug discovery papers containing experimental validation.
To ensure a fair and meaningful comparison across these papers, we applied filters to the data extracted from each study. We focused exclusively on compounds that could be classified under the Hit Identification campaign, as this represents the first and most challenging application in early-stage AI-driven drug discovery. Additionally, campaigns were limited to those where at least ten compounds were screened in vitro per target, creating a more robust statistical framework based on the hypothesis that AI models should be reproducible and capable of discovering entire libraries of novel hits, rather than isolated successes. We then ensured that only the exact compounds predicted by the AI models—without any intermediate or high-similarity synthetically accessible alternatives—were considered in the analysis. This allows us to truly evaluate the AI model’s performance by eliminating any random chance of success, ensuring that only the compounds directly predicted by the AI are credited as hits. Furthermore, for a compound to be classified as a hit, it needed to demonstrate not just binding affinity (Kd) but also biological activity against the target protein at or below a concentration of 20 μM. Non-standardized concentrations used to define “hits” can lead to inflated hit rates by permitting concentrations that are too high to be therapeutically viable. While some argue that a lower cutoff, such as 1 μM, is more appropriate for therapeutic viability, our focus during the Hit Identification stage is on identifying compounds with activity levels within an acceptable range for therapeutic-level optimization. At this early stage of development, we can be somewhat more permissive with the cutoff because the goal is to discover novel SAR that can be further optimized for potency in later stages. By setting a 20 μM threshold, we ensure that we capture compounds with genuine potential, which can then be refined and enhanced as they progress in development. The results of these filters can be seen in Table 2.
Table 2.
Adjusted hit rates from a review of studies [5] and additional sources based on preliminary filters.
a. The data includes hits at concentrations up to 250 µM, with some compounds acting only as binders and could not be further decomposed.
b. The data includes hits at concentrations up to 30 µM and could not be further decomposed.
Hit rates reported by Atomwise and Schrödinger, as displayed in Table 2, were excluded from our analysis beyond this point. This decision was made because their data could not be further decomposed based on the information provided, limiting our ability to conduct a proper assessment of their hit rates.
To continue our analysis of AI-driven Hit Identification campaigns, we assessed chemical novelty using Tanimoto similarity applied to ECFP4 2048-bit encodings of molecules**. Tanimoto similarity, a widely used industry metric, measures chemical similarity by quantifying the shared structural components between two molecules on a scale from 0 to 1, where 0 indicates no similarity and 1 indicates identical compounds. In this early phase of chemical discovery, the identified hits must be chemically distinct from known compounds against the target protein and also diverse among themselves. We approached this assessment in three ways, each serving a different purpose. First, we assessed the chemical similarity of the hits relative to the compounds used to train the model, directly testing the model’s capacity to predict beyond the chemical space it was trained on. Second, we evaluated the chemical similarity of the discovered hits compared to all known compounds with bioactivity against the same target protein at the time of discovery, providing insight into the general novelty of the compounds. These first two similarities were calculated using Average Nearest Neighbor (or Average Maximum) Tanimoto similarity between the hits and the comparison set. Lastly, we calculated pairwise similarity scores among the hits to evaluate their diversity, ensuring that the hits represent a wide range of chemical structures. The pairwise diversity score was calculated as the average Tanimoto similarity across all possible pairs of hits, excluding self-similarity. By applying these standardized criteria, we aimed to create a level playing field that accurately reflects the capabilities of AI models in identifying truly novel and active compounds.
Footnote: **We applied consistent methods for data collection and analysis across all studies. When available, we incorporated the training data provided in each publication into our analysis. To compile a comprehensive set of all known compounds with bioactivity against the same target protein at the time of discovery, we relied on the ChEMBL database, selecting versions released before the discovery dates disclosed in the publications.
Table 3.
Adjusted hit rates from review article [5] and additional sources based on preliminary filters and assessment of chemical novelty.
a. No training set available.
b. No hits to run analysis on after filters were applied.
c. Only one compound in the set of hits, thus unable to calculate Pairwise Diversity Score.
As seen in Table 3, most models struggle to demonstrate significant chemical novelty in their discovered hits and show a lack of diversity amongst those hits, highlighting their limitations in predicting the properties of compounds beyond their trained chemical space. This challenge becomes particularly clear when Tanimoto coefficients drop below the industry standard of 0.5, which is commonly used to assess chemical novelty [16]. For instance, the LSTM RNN model [6], while achieving a 43% hit rate, shows a high Tanimoto similarity of 0.66 to both the training data and the ChEMBL database, indicating that it is largely rediscovering known chemistry rather than breaking into new chemical territory. The GRU RNN model [7] claims a 88% hit rate, however, the lack of available training set data makes it difficult to fully assess the novelty of its discoveries and its pairwise diversity of 0.28 score indicates a lack of diversity within its hits. Similarly, the Stack-GRU RNN model [10] shows a hit rate of 27%, but Tanimoto similarities of 0.49 and 0.55 to the training and ChEMBL data, respectively, suggest it is also rediscovering known chemical structures rather than identifying truly novel compounds. These new data insights provide a deeper understanding of each model's capabilities, revealing that even models with claimed high hit rates still fall short in exploring novel chemical space and producing diverse, innovative compounds.
In contrast, our recent paper, ChemPrint: An AI-Driven Framework for Enhanced Drug Discovery [4], validates ChemPrint’s superior performance in two distinct campaigns targeting the oncoproteins AXL and BRD4. For AXL, ChemPrint achieved a 41% hit rate (12 out of 29), with a Tanimoto similarity score of 0.4 to both the training data and the ChEMBL database, indicating a further exploration of novel chemical space. For BRD4, ChemPrint demonstrated an even more impressive 58% hit rate (7 out of 12), with Tanimoto similarity scores of 0.3 to the training data and 0.31 to the ChEMBL database. Additionally, ChemPrint’s pairwise diversity scores were 0.17 for AXL and 0.11 for BRD4, reflecting diversity among the hits, outperforming other models (Table 3). When models are evaluated using rigorous and appropriate criteria, ChemPrint stands out as the leading performer. The reasons behind ChemPrint's success and an explanation of approach can be found in our recent ChemPrint paper [4]. We recognize that the difficulty of target druggability also plays a role and can impact hit rates. Even though we did not normalize hit rates to protein target druggability, our analysis is still impactful as AXL and BRD4 are notoriously hard-to-drug targets, thus the ChemPrint data used here represents a likely worst-case, case-study.
To provide a framework for assessing the capability of AI models in this space moving forward, it is essential to consider the rigorous standards we applied in our analysis. We used our evaluation guidelines, specifically designed for Hit Identification campaigns, to evaluate all models fairly and consistently. This ensures that only the most robust predictions are recognized by setting stringent criteria for in vitro testing, chemical novelty, and diversity. These guidelines should serve as a marker of progress, helping to benchmark and drive the development of future AI-driven drug discovery initiatives.
Evaluation Guidelines for AI Hit Identification Models:
Minimum Sample Size: A minimum of 10 compounds must be tested in vitro, ensuring that the analysis is based on a robust sample size. This criterion allows for meaningful comparison across studies and reflects the expectation that AI models should be capable of discovering entire libraries of hits, rather than isolated successes.
Exact Compound Prediction Testing: Only the exact compounds predicted by the AI models are tested, without any intermediate or high-similarity synthetically accessible alternatives. This eliminates any random chance of success, ensuring that only the compounds directly predicted by the AI are credited as hits.
Therapeutic Relevance: The compound must demonstrate not just binding affinity (Kd) but also biological activity against the target protein at or below a concentration of 20 μM. This cutoff ensures that only compounds with genuine potential, and that elicit a meaningful biological response, are considered hits, avoiding inflated hit rates from non-therapeutic concentrations.
Chemical Novelty: An average nearest neighbor Tanimoto similarity score (ECFP4 2048) to the training data must be less than 0.4, to extend the novelty of the chemical space being explored.
Public Data Novelty: An average nearest neighbor Tanimoto similarity score (ECFP4 2048) to public data must also be less than 0.4, further reinforcing the discovery of novel compounds not already known in the broader scientific community.
Diversity Among Hits: An average pairwise Tanimoto diversity score (ECFP4 2048) must be less than 0.2, ensuring diversity among the identified hits and confirming that the AI model is capable of exploring a broad range of chemical structures.
In conclusion, our analysis of the landscape of AI-driven small molecule drug discovery highlights the importance of rigorous evaluation criteria, especially within Hit Identification campaigns, where the challenge of discovering novel compounds is most pronounced. By applying standards for in vitro validation, chemical novelty, and diversity, we have demonstrated that even models with high reported hit rates can fall short in exploring novel chemical spaces. ChemPrint, however, has distinguished itself by demonstrating superior hit rates while also identifying highly novel and diverse chemical compounds. This emphasizes the need for the industry to adopt robust benchmarking frameworks, such as our evaluation guidelines, to ensure meaningful comparisons and advancements in AI-driven drug discovery.
FIGURE 1
We evaluated the models using a comprehensive ranking system based on four key metrics: Hit Rate, Tanimoto Similarity to Training Data and ChEMBL Data, and Pairwise Diversity Score. Models were assigned points for their performance in each category (1 for best, 2 for second-best, etc.), with missing data assigned the average of remaining ranks. Total points were calculated by summing across all metrics, with lower scores indicating better overall performance. This approach provides a balanced evaluation, equally weighting each metric to offer a holistic view of each model's effectiveness in both identifying active compounds and exploring novel chemical space.
References
(1) Lloyd, M. D. High-Throughput Screening for the Discovery of Enzyme Inhibitors. Journal of Medicinal Chemistry, 2020, 63, 10742–10772. https://doi.org/10.1021/ acs.jmedchem.0c00523.
(2) Schrödinger, Inc. Dramatically Improving Hit Rates with a Modern Virtual Screening Workflow. Schrödinger: Life Sciences. https://www.schrodinger.com/life-science/learn/white-papers/dramatically-improving-hit-rates-modern-virtual-screening-workflow/ (accessed 2024-04-18).
(3) Insilico Medicine. Precious3GPT: Accelerating Drug Discovery with AI. Insilico Medicine Repository. https://insilico.com/repository/precious3gpt (accessed 2024-07-31).
(4) Umansky, T. J., Woods, V. A., Russell, S. M., Smith, D. M., & Haders, D. J. (2024). ChemPrint: An AI-Driven Framework for Enhanced Drug Discovery. bioRxiv. https://doi.org/10.1101/2024.03.22.586314
(5) Du, Y.; Jamasb, A. R.; Guo, J.; Fu, T.; Harris, C.; Wang, Y.; Duan, C.; Liò, P.; Schwaller, P.; Blundell, T. L. Machine Learning-Aided Generative Molecular Design. Nature Machine Intelligence, 2024, 6, 589–604. https://doi.org/10.1038/s42256-024-00843-5 .
(6) Grisoni, F., Huisman, B. J. H., Button, A. L., Moret, M., Atz, K., Merk, D., & Schneider, G. (2021). Combining generative artificial intelligence and on-chip synthesis for de novo drug design. In Science Advances (Vol. 7, Issue 24). American Association for the Advancement of Science (AAAS). https://doi.org/10.1126/sciadv.abg3338
(7) Hua, Y., Fang, X., Xing, G., Xu, Y., Liang, L., Deng, C., Dai, X., Liu, H., Lu, T., Zhang, Y., & Chen, Y. (2022). Effective Reaction-Based De Novo Strategy for Kinase Targets: A Case Study on MERTK Inhibitors. In Journal of Chemical Information and Modeling (Vol. 62, Issue 7, pp. 1654–1668). American Chemical Society (ACS). https://doi.org/10.1021/acs.jcim.2c00068
(8) Moret, M., Pachon Angona, I., Cotos, L., Yan, S., Atz, K., Brunner, C., Baumgartner, M., Grisoni, F., & Schneider, G. (2023). Leveraging molecular structure and bioactivity with chemical language models for de novo drug design. In Nature Communications (Vol. 14, Issue 1). Springer Science and Business Media LLC. https://doi.org/10.1038/s41467-022-35692-6
(9) Yu, Y., Huang, J., He, H., Han, J., Ye, G., Xu, T., Sun, X., Chen, X., Ren, X., Li, C., Li, H., Huang, W., Liu, Y., Wang, X., Gao, Y., Cheng, N., Guo, N., Chen, X., Feng, J., … Li, H. (2023). Accelerated Discovery of Macrocyclic CDK2 Inhibitor QR-6401 by Generative Models and Structure-Based Drug Design. In ACS Medicinal Chemistry Letters (Vol. 14, Issue 3, pp. 297–304). American Chemical Society (ACS). https://doi.org/10.1021/acsmedchemlett.2c00515
(10) Korshunova, M., Huang, N., Capuzzi, S., Radchenko, D. S., Savych, O., Moroz, Y. S., Wells, C. I., Willson, T. M., Tropsha, A., & Isayev, O. (2022). Generative and reinforcement learning approaches for the automated de novo design of bioactive compounds. In Communications Chemistry (Vol. 5, Issue 1). Springer Science and Business Media LLC. https://doi.org/10.1038/s42004-022-00733-0
(11) Vakili, M. G., Gorgulla, C., Nigam, A., Bezrukov, D., Varoli, D., Aliper, A., Polykovsky, D., Das, K. M. P., Snider, J., Lyakisheva, A., Mansob, A. H., Yao, Z., Bitar, L., Radchenko, E., Ding, X., Liu, J., Meng, F., Ren, F., Cao, Y., … Zhavoronkov, A. (2024). Quantum Computing-Enhanced Algorithm Unveils Novel Inhibitors for KRAS (Version 1). arXiv. https://doi.org/10.48550/ARXIV.2402.08210
(12) Swanson, K., Liu, G., Catacutan, D. B., Arnold, A., Zou, J., & Stokes, J. M. (2024). Generative AI for designing and validating easily synthesizable and structurally novel antibiotics. In Nature Machine Intelligence (Vol. 6, Issue 3, pp. 338–353). Springer Science and Business Media LLC. https://doi.org/10.1038/s42256-024-00809-7
(13) Ren, F., Ding, X., Zheng, M., Korzinkin, M., Cai, X., Zhu, W., Mantsyzov, A., Aliper, A., Aladinskiy, V., Cao, Z., Kong, S., Long, X., Man Liu, B. H., Liu, Y., Naumov, V., Shneyderman, A., Ozerov, I. V., Wang, J., Pun, F. W., … Zhavoronkov, A. (2023). AlphaFold accelerates artificial intelligence powered drug discovery: efficient discovery of a novel CDK20 small molecule inhibitor. In Chemical Science (Vol. 14, Issue 6, pp. 1443–1452). Royal Society of Chemistry (RSC). https://doi.org/10.1039/d2sc05709c
(14) Eguida, M., Schmitt-Valencia, C., Hibert, M., Villa, P., & Rognan, D. (2022). Target-Focused Library Design by Pocket-Applied Computer Vision and Fragment Deep Generative Linking. In Journal of Medicinal Chemistry (Vol. 65, Issue 20, pp. 13771–13783). American Chemical Society (ACS). https://doi.org/10.1021/acs.jmedchem.2c00931
(15) Wallach, I., Bernard, D., Nguyen, K., Ho, G., Morrison, A., Stecula, A., Rosnik, A., O’Sullivan, A. M., Davtyan, A., Samudio, B., Thomas, B., Worley, B., Butler, B., Laggner, C., Thayer, D., Moharreri, E., Friedland, G., Truong, H., … Heifets, A. (2024). AI is a viable alternative to high throughput screening: a 318-target study. In Scientific Reports (Vol. 14, Issue 1). Springer Science and Business Media LLC. https://doi.org/10.1038/s41598-024-54655-z
(16) Hu, Y., Stumpfe, D., & Bajorath, J. (2013). Advancing the activity cliff concept. In F1000Research (Vol. 2, p. 199). F1000 Research Ltd. https://doi.org/10.12688/f1000research.2-199.v1
Written by Tyler Umansky